
ДНК-нанотехнологии 1 введение и основные методы / 10.1021@acs.chemrev.0c00294
.pdf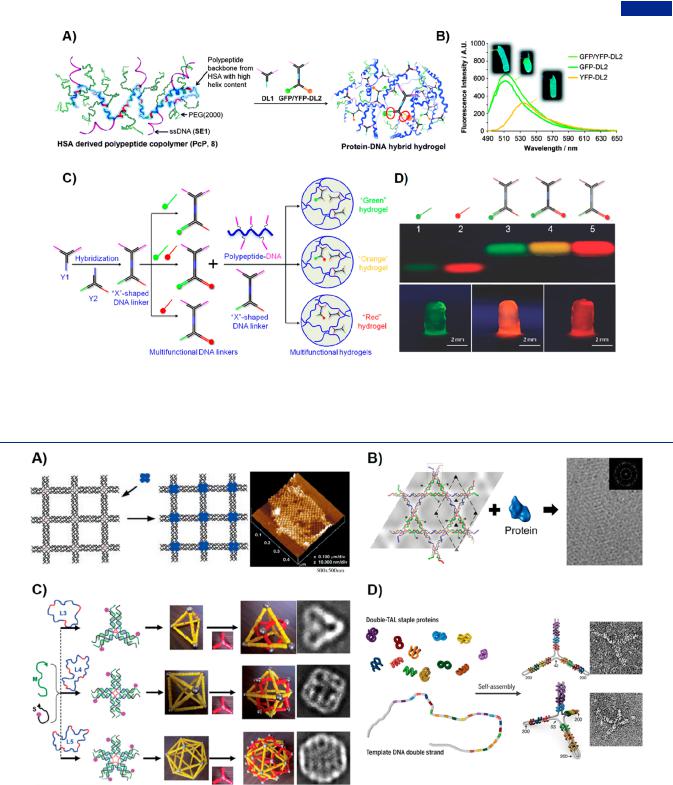
Chemical Reviews |
pubs.acs.org/CR |
Review |
Figure 33. Branched DNA-proteins hybrids. (A) Synthesis of a protein-DNA hybrid hydrogel from polypeptide copolymers and branched DNA. (B) Fluorescence properties of protein-DNA hybrid hydrogels containing green fluorescent protein (GFP) and yellow fluorescent protein (YFP). Adapted with permission from ref 354. Copyright 2014, Royal Society of Chemistry. (C) Synthesis of multifunctional protein-DNA hydrogels composed of X- shaped DNA linker and polypeptide-DNA. (D) Tunable fluorescence properties of protein-DNA hybrid hydrogels by integrating GFP and YFP. Adapted with permission from ref 355. Copyright 2014, Wiley-VCH.
Figure 34. Branched DNA-templated addressable distribution of protein. (A) 2D DNA arrays-templated self-assembly of proteins and its AFM image. Adapted with permission from ref 124. Copyright 2003, American Association for the Advancement of Science. (B) Single-particle cryoEM for characterizing the guanine nucleotide-binding protein (Gαi1) bound to DNA array template. Adapted with permission from ref 105. Copyright 2011, American Chemical Society. (C) Various DNA polyhedron-directed 3D organization of protein by the specific binding of biotin-modified DNA nanostructures and streptavidin (STV) protein. Corresponding 2D projections of reconstructed models were adapted with permission from ref 360. Copyright 2012, Wiley-VCH. (D) A set of custom proteins (transcription activator-like e ector proteins, TAL proteins) acted as staples to control the spatial arrangement of user-defined branched DNA-protein hybrids. On the right were the TEM micrographs of single three-armed branched DNAprotein hybrid and four-armed branched DNA-protein hybrid. Adapted with permission from ref 363. Copyright 2017, American Association for the Advancement of Science.
AE |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. XXXX, XXX, XXX−XXX |

Chemical Reviews |
pubs.acs.org/CR |
Review |
strands could be synthesized as triblock DNA-ligands conjugates, which assembled into a triangular face with singlestranded linkers. The proximity of ligands facilitated the selective metal coordination of two triangular faces at vertices, eventually leading to the accessibility of metalated DNA cages (Figure 32B).85 The addressability of branched DNA-based nanocages conducted to accomplish the programmable positioning of transition metal by guest-mediated assembly. In addition, the mediation of transition metal−ligands was accessible to the formation of chiral branched DNA with four di erent arms, which was considered as template to guide the
assembly of DNA nanotubes by the assistant of single strands (Figure 32C).350
3.4.4. DNA-Peptide/Protein Hybrids. Peptides and proteins possessed unique amino acid sequences, well-defined secondary structures as well as many functional groups that were easily chemically modified, therefore allowing functional proteins to be attached to DNA by covalent chemical reactions or noncovalent physical interactions.351−353 For example, Wu et al. reported a protein-DNA hybrid hydrogel composed of modified polypeptide copolymers as backbones and four-arm DNA labeled with fluorescent proteins as cross-linkers (Figure 33A).354 The protein backbones with azido groups were conjugated to alkyne-modified DNA via click reaction, resulting in DNA-polypeptide copolymers. Afterward, the functionalized multiarm DNA was designed as cross-linkers to connect copolymers for the construction of protein-DNA hybrid hydrogels. The functionalized multiarm DNA was formed by the self-assembly of Y-DNA building-blocks, and then modified with GFP and yellow fluorescent protein (YFP) simultaneously though the site-specifically conjugation between maleimidemodified DNA and single-mutated cysteine residues. The introduction of GFP and YFP endowed the protein-DNA hybrid hydrogels with fluorescence (Figure 33B). Employing the same method of ligation between protein and DNA, Gacaniň et al. prepared a multifunctional DNA-protein hybrid hydrogel with the implant of therapeutically active molecules, recombinant Rho-inhibiting C3 toxin, for the specially target of osteoclasts, inhibition of Rho-signaling pathway and further local treatment of a kind of bone diseases, osteoporosis.310 Liu group fabricated supramolecular hydrogels based on the simple mixing of DNA-grafted polypeptide and X-shaped DNA linkers with multiple functionalization sites (Figure 33C).355 These hydrogels showed di erent colors by regulating the fluorophores on functional sites (Figure 33D). Furthermore, the hydrogels possessed excellent self-healing and rapid-responsive thixotropic properties, enabling the direct-writing of arbitrary 3D structures. In addition, protein served as sca olds and branched points for the construction of DNA hybrid nanohydrogels via the high- a nity interaction between streptavidin (SA)-modified branched DNA and biotin.356 The size of DNA nanohydrogel was easily controlled by tuning the concentration of branched DNA motif. Owing to the incorporation of fluorophores, ATP aptamer, tumor-targeted aptamer and therapeutic molecules, the protein-sca olded DNA nanohydrogel was capable of synergistic functionalities of targeted delivery, ATP-activated imaging, drug release and cancer therapy.
The precise addressability of regular DNA nanostructures (such as 2D array and 3D polyhedron) has made deterministic positioning of protein in space possible.357,358 Yan et al. constructed periodic protein arrays templated by well-designed DNA patterns self-assembled from four-armed branched DNA.124 The DNA patterns were designed as two distinct
lattice morphologies, namely uniform-width nanoribbons and 2D nanogrids. The DNA nanogrids as templates were functionalized with biotin groups in the large cavities for the periodic binding of SA proteins (Figure 34A). The DNAtemplated protein arrays showed dense and nonoverlapping distribution, therefore facilitating high-resolution electron cryomicroscopy at the single-molecule level (Figure 34B).105 In addition, 3D DNA nanostructures provided addressing sites to precisely control the spatial distribution of proteins. Proteins could attach on 3D DNA nanostructures by covalently linking of biotin-containing bifunctional linker and phosphorothioate (PS) backbone-modified DNA as anchor strands where phosphate backbones of thymines at the polyhedral verse were replaced by PS. The resulting biotin-linker-modified DNA polyhedron was incubated with SA proteins to obtain di erent amounts of SA-decorated DNA polyhedrons.359 Because SA protein had four binding sites with biotin, this monovalent biotin-modified DNA polyhedron cannot precisely control the amount and positions of SA proteins binding. To control the freedom of bound SA, a series of DNA polyhedrons with trivalent biotin modified on each face were developed (Figure 34C).360 The trivalent binding was attributed to the unique 3- fold rotational symmetry structure of DNA polyhedron, meanwhile improving the binding strong relative to single biotin binding, and e ciently controlling the guest freedom to avoid unexpected binding. This DNA sca old-based assembly strategy was also applicable to the orientated assembly of other proteins. In addition to using DNA as templates to prepare ordered protein assemblies, proteins in turn can promote welldefined DNA assembly.361 For example, the incorporation of certain bacterial recombination protein, RuvA, influenced the morphology, symmetry, and connectivity of DNA array, from
antiparallel x-stacked configuration (60° angles) to a squareplanar configuration (90° angles).362 Inspired by the DNA
origami strategy, Praetorius et al. proposed a bottom-up fabrication strategy to synthesize user-defined DNA−protein hybrids at the desired scale, including three-armed and fourarmed shapes, by using a set of custom “staple” proteins (Figure 34D).363 The transcription activator-like (TAL) e ector proteins recognized and folded the specific DNA sequences in double strand template to form a superhelix. To function as staple, two TAL protein were fused together to link two distinct DNA recognition domains, achieving the assembly of DNAprotein hybrids with custom spatial arrangement and geometries in a one-pot reaction.
4. APPLICATIONS
4.1. Diagnostics
Nucleic acid-based diagnostics have attracted ever-increasing attentions driven by the development of biotechnologies.364 By virtue of the specific base complementary pairing, DNA have been widely utilized for the detection of target nucleic acids (DNA/RNA) in the diagnosis of viral infections, bacterial infections, and cancer in recent decades.365−371 Meanwhile, the emergence of DNA aptamers that specifically bound with target with high a nity made the scope of detection analytes not limited to nucleic acids, but extended to proteins and other small molecules such as thrombin, ATP, antibiotics, and biotoxin.372−377 In addition, the coordination interaction of DNA with metals ions (such as T-Hg2+-T and C-Ag+-C complex) created possibilities for the sensing of hazardous heavy metal.378−380 It
AF |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. XXXX, XXX, XXX−XXX |
Chemical Reviews |
pubs.acs.org/CR |
Review |
Figure 35. Sandwich assay based on branched DNA as signal amplifiers. (A) Schematic representation of the construction of a recyclable biointerface based on single branched DNA for nucleic acid detection. (B) Frequency response curves showing two-stage decline by using chemically cross-linked branched DNA as probes and preamplifiers, and DNA functionalized Fe3O4 NPs as postamplifiers. Adapted with permission from ref 389. Copyright 2018, Elsevier Publishing Group. (C) Cascade-amplification detection platform based on two kinds of X-DNA as signal amplifiers.391 (D) Frequency response curves presenting stepped decreasing when alternately introducing two kinds of X-DNA. (E) Multistepped amplification platform e ectively distinguishing the di erent positions of single base mutation.
followed that DNA-based diagnostic methods played tremendous values in the detection of a large range of analytes.
Not only did branched DNA have the capability of accurate and rapid recognition with manifold target molecules but also its unique multiarmed structures could serve as a versatile signal amplifier for the sensitive, convenient, personalized, and multiplexed diagnosis.381−387 By coupling branched DNA and a variety of amplification strategies, a large number of branched DNA-based assays were developed. According to the diverse contributions of branched DNA structures in diagnostics, the diagnostics strategies were catalogized into sandwich-based assay (including one-step amplification and multistep amplifi- cation), target-induced polymerization, target recycling strategy, and multiplexed diagnostics.
4.1.1. Sandwich-Based Assay. Sandwich-based assay, as a powerful technique, has been extensively developed in the fields of nucleic acid diagnostics. In general, sandwich-based assay was compatible with a variety of biosensors, such as electrochemical, chemiluminescence, fluorescence, colorimetric, surface plasmon resonance, surface-enhanced Raman scattering (SERS), quartz crystal microbalance, microcantilever, and other biosensing devices, having the characteristics of simple design, convenient operation, and flexible employment.388
A typical sandwich-based assay consists of two parts: recognition probes for binding analytes; signal molecules for amplification. Branched DNA could be used as recognition
probes and signal amplifiers simultaneously in sandwich-based strategies. Using the typical “sandwich” model, Li et al. created a branched DNA-based, recyclable biointerface for nucleic acid detection (Figure 35A).389 In this system, chemically crosslinked branched DNA nanostructures were utilized as probes, and DNA functionalized Fe3O4 nanoparticles were used for signal amplification (Figure 35B). The branched DNA probes not only enhanced the signal through increasing the distances among probes and decreasing space hindrance but also provided the regeneration of detection system by advantage of antidenaturability of chemically cross-linked branched DNA. The recyclable biointerface achieved a limit of detection (LOD) as low as 500 fM. By combing the antigen−antibody recognition on a single platform, Cheng et al. designed an ultrasensitive SERS biosensor based on branched DNA-assisted sandwich strategy for the simultaneous detection of miRNA and protein.390 This sensing platform exploited Y-branched DNA as probe for capturing miR-223 and antibody for specifically binding to α-fetoprotein, showing extremely low limit of detection as 10−17 M and 10−12 M, respectively. Worthy to note was that the implantation of branched DNA probes enabled 2 orders of magnitude increase compared with single-stranded DNA. This system was highly amenable to the detection of miRNA and protein in complex physiological media, exhibiting great application prospects for early diagnosis of primary cancers.
AG |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. XXXX, XXX, XXX−XXX |
Chemical Reviews |
pubs.acs.org/CR |
Review |
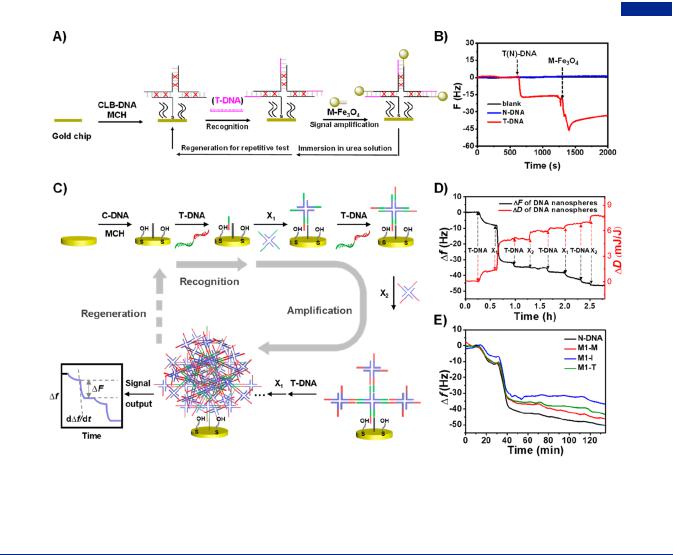
Figure 36. Branched DNA-based nucleic acid detection by the strategy of target-induced polymerization. (A) Schematic of target-driven polymerization for the detection of pathogen target DNA. (B) Quantum dots-encoded ABC monomers polymerizing into nanospheres, showing orange fluorescence in the presence of pathogen DNA. Scale bar, 5 μm. (C) Fluorescence microscopy image of polymeric nanospheres in Hela cells. Scale bar, 10 μm. Adapted with permission from ref 74. Copyright 2009, Springer Nature.
Relative to single branched DNA nanostructure, DNA dendrimers possessed highly branched architecture and larger molecular weight, such that DNA dendrimers functioned as enhanced signal amplifiers in the presence of equivalent analytes. In addition, DNA dendrimers had larger surface area and abundant binding sites for the high loading capacity of signal molecules, which made DNA dendrimers more promising signal
nanocarriers and amplifiers in sandwich-based biosensors.392−394 By the aid of layer-by-layer assembled DNA
dendrimer, Wang et al. constructed a electrochemiluminescence (ECL) biosensor for sensitive detection of glucosaminidase by embedding luminescent Ru(II) complex into DNA dendrimers, in which the DNA dendrimers provided abundant intercalation sites of Ru(II) complex for enhancing luminescent e ciency.395 Similarly, Li et al. proposed a ECL immunosensor based on DNA dendrimers as nanocarriers for ultrasensitive detection of protein.396 The DNA dendrimers were formed by the step-by- step self-assembly between two types of chemically synthesized Y-shaped DNA, sense Y-DNA, and its antisense Y-DNA. After the intercalation with luminescent indicators, DNA dendrimers served as amplification probes for the ultrasensitive detection of target protein. Additionally, DNA dendrimers could also be prepared by dynamic assembly triggered by initiators. Liu et al. constructed an enzyme-free and label-free electrochemical biosensor for the detection of DNA.397 The DNA dendrimers were fabricated by three auxiliary DNA chains and served as signal output and amplification molecules due to abundant binding sites for the intercalation of electroactive molecules. The DNA dendrimer-based electrochemical biosensor demonstrated significantly enhanced amplification performance and better sensitivity (5 aM) compared with linear DNA nanostructures. On the basis of dynamic-assembled DNA dendrimers, Zhao et al. incorporated SA proteins and DNA linker to prepare DNA dendrimers-SA nanocomplex for the construction of signal amplification platform.254 The DNA dendrimers-SA nanocomplex could realize the sensing of disease-related gene fragments and cancer cells with the aid of mass-sensitive piezoelectric sensors and fluorescence technology, respectively.
Li et al. assembled DNA dendrimers-Au nanoaggregates by the dynamic assembly of three species of hairpin DNA.323 The participation of DNA dendrimers made the nanoaggregates with Raman dye molecules have a great quantity of SERS “hot spots”, which functioned as probes and signal amplifiers to achieve simultaneous detection of two biomarkers on the cancer cells, and SERS-based cell imaging.
On the basis of one-step amplification, a strategy of multistep amplification has been developed to further improve detection
performance. Dong et al. developed a strategy of step-by-step amplification based on branched DNA (Figure 35C).391 In the
presence of target DNA, X-shaped DNA specifically and rapidly polymerized into DNA dendrimers without the involvement of any enzymes. By alternatively introducing two kinds of X-DNA, the frequency signals and corresponding dissipative signals presented a trend of stepwise change for enhanced detection sensitivity (Figure 35D). The system exhibited robust and reliable in a wide range of harsh environments. Specially, this system had the capability of discriminating mutated DNA from normal DNA, including 1−4 mutated bases and di erent positions of single base mutations (Figure 35E). Making use of multistep polymerization mechanism, Zhao et al. introduced SA into DNA dendrimer as a new signal amplifier for the
construction of quartz crystal microbalance (QCM) biosensing platform.254,398 Because DNA dendrimer-SA was formed by the
trigger of target and the cascaded assembly, the platform exhibited gradual signal trends when alternately introducing two branched DNA-SA complexes as signal amplifiers. The DNA dendrimer-SA-based detection platform showed excellent selectivity and sensitivity for detecting disease-related gene fragment and discriminating single-base mutation. Guo et al. encapsulated SA into layer-by-layer assembled DNA hydrogels using the specific recognition and binding of aptamers and SA.399 The constructed surface plasmon resonance (SPR) biosensor based on SA-DNA hydrogels as supere cient amplifiers was developed for ultrasensitive detection of fusion gene. The characteristics of step-by-step amplification mechanism for assembling DNA nanostructures provided an
AH |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. XXXX, XXX, XXX−XXX |

Chemical Reviews |
pubs.acs.org/CR |
Review |
opportunity for superior specificity in nucleic acid detection, especially single nucleotide mutations.
Branched DNA assay, as a classic sandwich-based assay, was based on a series of hybridization reactions, which achieved quantitative detection of target nucleic acids in terms of early disease diagnosing.400 This technology included immobilized capture probes, capture extenders for specially hybridizing to capture probes and target sequences, multistage branched DNA amplifier molecules, and chemiluminescent label extenders to signal output.401 It is noteworthy that the technique did not require the participation of enzymes and rigorous experimental conditions. By virtue of the technology, Liu et al. introduced SAcoated magnetic beads as fixed substrates and Ru(bpy)32+- labeled probes in replace of traditional alkaline phosphatase labels, consequently achieving the ultrasensitive electrochemiluminescence detection of target DNA in pathogenic bacteria, Staphylococcus aureus.371,402 In addition, this method could obtain the measurement results of mRNA concentration similar to traditional real-time quantitative PCR (RT-qPCR) approach, and was a sensitive, accurate, less-laborious, and automated tool alternative to the RT-qPCR.403 The PCR-free branched DNA assay therefore was conducive to the quantitative detection of target transcripts in mRNA population for early disease markers.404
4.1.2. Target-Induced Polymerization. A new mechanism called “target-induced polymerization” was first proposed by Luo group,74 in which branched DNA could polymerize into larger aggregation induced by target molecules. Target molecules acted as not only the linkers, but also the trigger chains to induce the polymerization of branched DNA, ensuring the specificity and sensitivity of reaction. Moreover, the targetinduced polymerization reaction occurred without the participation of enzymes and other biosensing devices. Specially, they first designed a kind of anisotropic, branched and cross-linking building-blocks (ABC monomers) assembled from branched DNA. The resulting ABC monomers were constituted by X- DNA acceptors and Y-DNA donors with multiple and varying functional moieties, including quantum dots, gold nanoparticles, fluorescent dyes and photo-cross-linking groups. To achieve enzyme-free pathogen DNA diagnostics, the ABC monomers were customed with two kinds of quantum dots (red and green colors), one photo-cross-linking group, and one ssDNA probe that was complementary to a specific pathogen DNA (Figure 36A). With the promotion of target pathogen DNA, two monomers self-assembled to form a dimer. Afterward, the resultant dimers aggregated into a sphere upon exposure to UV illumination, which showed unique color codes in both bright filed and fluorescent field. Remarkably, when the ratio of green quantum dots and red quantum dots on dimers was 1:1, the polymeric nanospheres exhibited orange fluorescence (Figure 36B). This target-induced polymerization of ABC monomers greatly amplified the signal of pathogen DNA, resulting in rapid detection with high specificity and sensitivity in living cells (Figure 36C). The construction of multifunctional nanoarchitectures from monomers provided a versatile route for the applications in nanoelectronics, nanophotonics, intelligent sensing, and drug delivery.
Alternatively, dynamic formation of DNA dendrimers only triggered by target was another manifestation of target-induced polymerization. Xuan et al. took advantage of the dynamic assembly of DNA dendrimers to develop an enzyme-free and immobilization-free electrochemical nucleic acid sensing method.405 A kind of nucleic acid analogue-peptide nucleic
acid labeled by ferrocene (Fc-PNAs) was incorporated into DNA sequences, and autonomously assembled into DNA dendrimers in a kinetically controlled fashion. In the absence of target, the components maintained structural stability, and freely di usible Fc-PNAs produced electrochemical signal. With the addition of target nucleic acid, the dynamic assembly occurred in situ on the electrode surface. Because of the successive consumption of Fc-PNAs and strong electrostatic repulsion between DNA/Fc-PNAs dendrimers and electrodes with identical negatively charged, the electrochemical signal appeared significant reduction. Thus, the sensing method achieved high-sensitive and high-specific nucleic acid detection. Zhou et al. proposed a method of coupling Y-shaped DNA with DNA dendrimers constructed from dynamic assembly strategy.406 When target microRNA (miRNA) was present, the competition between Y-shaped DNA and miRNA caused Y- shaped DNA to be degraded and initiation chains to be exposed,
finally yielding DNA dendrimers via a series of strand displacement reactions for remarkable electrochemical signal changes. In addition, target protein could also be designed as a promotor to activate the dynamic formation of DNA dendrimers.407 When target protein existed, hairpin structure with corresponding aptamer deformed and exposed to initiators to induce the formation of DNA dendrimers with the aid of strand displacement reaction-based dynamic assembly. DNA dendrimers constructed by target-induced polymerization could participate in the AuNPs-based colorimetric assay without sophisticated instrumentation. When unmodified AuNPs served as signal indicators, the formation of DNA dendrimers drove the aggregation of unmodified AuNPs owing to substantial reduction of free DNA strands for stabilization. On the contrary, when AuNPs were design to attach with DNA initiators, DNA dendrimers in situ growth on the surface of AuNPs provided
greater spatial hindrance and further prevent the aggregation of AuNPs.408
The simplest process of target-induced polymerization was that target molecule acted as a component chain or trigger component to drive the formation or dissociation of branched DNA, which was used to quantitatively analyze target.384 For example, the component strands of branched DNA were labeled with fluorophores and corresponding quenchers. The presence of target molecules caused the conformational transition of branched DNA, resulting in the dramatic change of signal intensity based on FRET e ect.386,410 The sensing systems could be integrated into DNA dendrimers-based systems to facilitate in situ imaging of intracellular biological molecules.411 The mechanism of target-induced polymerization could also be applied in the aggregation of DNA-capped AuNPs in SERS bioanalysis. Zhou et al. reported a target-induced assembly strategy of AuNPs dimers for SERS imaging of miRNAs (Figure 37A).409 On the basis of the electromagnetic theories, the nanoscale gap between AuNPs attributed to the generation of SERS “hot spots”, which led to greatly enhanced signals.412,413 The ingenious use of Y-shaped DNA e ectively narrowed the distance between AuNPs, resulting in in situ AuNPs dimers with high electromagnetic hot spots. A dramatically enhanced Raman signal therefore was obtained and presented on 3D Raman mapping images to monitor the spatial distribution of multiple miRNAs (Figure 37B). In addition, target-induced AuNPs aggregation were widely used in colorimetric technology due to intrinsic surface plasmon resonance e ect. AuNPs were easily aggregated triggered by salts in the form of bare gold, while maintained isolated when AuNPs were covered with a dense
AI |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. XXXX, XXX, XXX−XXX |

Chemical Reviews |
pubs.acs.org/CR |
Review |
Figure 37. Target-induced polymerization for the intracellular miRNAs imaging by the means of surface-enhanced Raman scattering. (A) Target-programmed Y-DNA-AuNPs dimers for enhanced Raman signals. (B) SERS images and corresponding signal intensities of miRNA in single cell after the treatment with Y-DNA-AuNPs dimers probes and two individual probes. Adapted with permission from ref 409. Copyright 2017, American Chemical Society.
DNA layer. Two DNA strands with perfect complementary to target could be designed to separately wrap on the surface of
AuNPs, and self-assembled into Y-DNA nanostructures and AuNP dimers with the trigger of target DNA, accompanied with
acolor change as naked-eyes output.383
4.1.3.Target Recycling Strategy. Target recycling
strategy evolved from the strategy of target-induced polymerization, and further improved the utilization of target. Target recycling could be essentially accomplished by two methods: kinetically controlled self-assembled process and enzymeassisted reactions.
Kinetically controlled self-assembled process was based on the CHA reaction. Target nucleic acid served as initiators to induce the beginning of CHA reaction and was then released for the next cycle, finally resulting in the formation of branched DNA or DNA dendrimers for improved signals. Wen et al. constructed a target-driven, nonenzymatic, and label-free sensing platform based on kinetically controlled self-assembly and AuNP colorimetric technique.414 The diblock DNA sequences were designed as hairpin block and anchoring block (polyA tails), which remained as hairpin conformation when in the absence of target, or converted to branched DNA induced by target DNA (Figure 38A). Hairpin DNA containing polyA tails that covered the surface of AuNPs could prevent the AuNPs aggregation induced by salt; while branched DNA transformed from hairpin DNA could reduce the distance between AuNPs, causing color changes (Figure 38B). Because of the recycling of target DNA, the platform could prominently improve the colorimetric signal sensitivity of target DNA. In addition, branched DNA constructed by CHA reaction could selfassemble to DNA dendrimers. The formation of DNA dendrimers made the hairpin DNA wrapped on the surface of AuNPs decrease dramatically, where hairpin DNA played a role in protecting AuNPs from aggregation. Thus, the obtained DNA
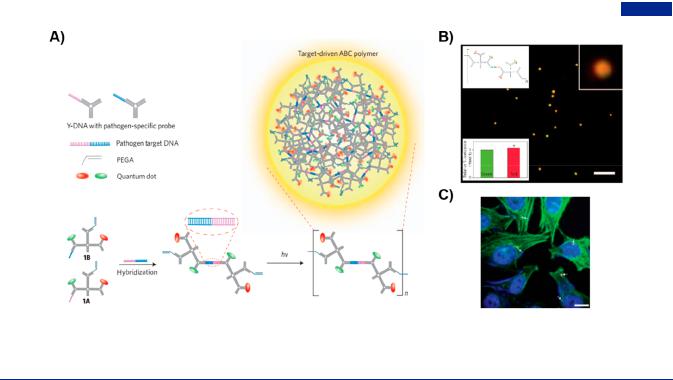
Figure 38. Branched DNA-based nucleic acid detection by advantage of target recycling strategy. (A) Construction of branched DNA with complementary sticky ends by target-triggered and target-recycled catalytic hairpin assembly (CHA) for the colorimetric detection of target DNA. (B) Colorimetric responses in the presence of target DNA with di erent concentrations. Adapted with permission from ref 414. Copyright 2016, Elsevier Publishing Group. (C) Intracellular RNA detection and in situ imaging based on branched DNA nanostructures. Illustration of the autonomous formation of Y-shaped DNA through target-induced catalysis assembly and polymerase-driven strand displacement amplification (SDA) reactions.
(D) Fluorescent imaging of in situ formation of Y-shaped DNA for intracellular miRNA diagnosis. On the right was the fluorescent intensity statistics of Y-shaped DNA formed by target-induced catalysis assembly. Adapted with permission from ref 369. Copyright 2018, Wiley-VCH.
AJ |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. XXXX, XXX, XXX−XXX |
Chemical Reviews |
pubs.acs.org/CR |
Review |
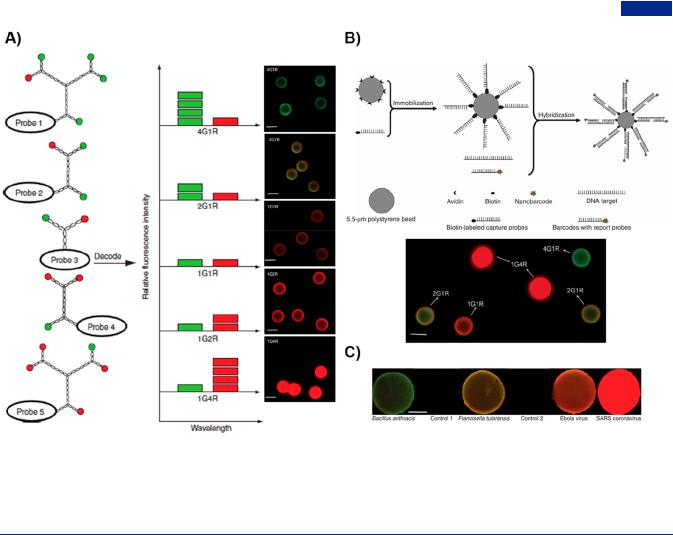
Figure 39. Branched DNA-based multiplexed detection of nucleic acid by nanobarcodes. (A) Scheme of Y-DNA-based nanobarcodes decoding with di erent ratio of fluorescence intensity and the detection of target DNA using nanobarcodes from microbeads (merged fluorescence images on the right). (B) Schematic representation of microbeads-based nanobarcodes for the multiplexed DNA detection by the strategy of sandwich assay. Multiplexed detection of target DNA using nanobarcodes with two-colored fluorescent groups (bottom). Scale bar, 5 μm. (C) Simultaneous detection of multiple pathogens DNA using a mixture of a series of nanobarcodes in a DNA blotting assay. Scale bar, 1 mm. Adapted with permission from ref 73. Copyright 2005, Springer Nature.
dendrimers also could be used to detect target nucleic acid in AuNPs-based colorimetric assay.132,252 In addition, target molecules in living cells could serve as catalysts to drive the formation of branched DNA-based functional materials, providing the opportunity for in situ intracellular imaging. Yue et al. developed a kind of target miRNA-catalyzed branched CHA reaction called bCHA for the diagnosis of intracellular miRNA.415 The target-catalyzed bCHA was considered as a versatile platform for in situ bioimaging and monitoring of miRNA. The enzyme-free catalytic assembly depended entirely on the expression level of target miRNA in live cells, exhibiting the detection specificity. Moreover, the amplification method was suitable for analysis of low-abundance miRNA by improving sensitivity. Bi et al. applied the target-catalyzed X-shaped DNA to intracellular imaging using chemiluminescence resonance energy transfer (CRET).416 A G-quadruplex DNAzyme embedded in X-DNA exhibited horseradish peroxidase-like activity when hemin/K+ was inserted. The DNAzyme could catalyze the production of luminol chemiluminescence and transfer to proximate fluorophore. In addition, the aptamerbased DNA nanohydrogels from X-DNA served as nanocarriers for bioimaging and drug delivery. Zhou and co-workers built a
target-triggered sensing system via CHA reactions for the detection of 17β-estradiol and bisphenol A.137,417 The aptamer
sequences were inserted on a duplex strands, and shielded the toehold domains from being recognized by other strands. Once target molecules were bound to aptamer sequences, the toehold domains were exposed and in turn triggered the CHA reaction
to form branched DNA. The sticky ends of branched DNA could be designed as hemin-inserted G-quadruplex DNAzyme with peroxidase-like activity, which catalyzed the color reactions to produce a visible signal in the presence of H2O2.137 Otherwise branched DNA could be intercalated with SYBR Green I, thus serving as fluorescent signal reporters.417 The label-free, targettriggered and recycling sensing platforms based on CHA reaction provide a cascaded signal amplification, and successfully realize the ultrasensitive and high-specific detection of target molecules.
Target recycling could also be accomplished through a variety of enzymes due to the ease of manipulation of DNA nanostructures, such as exonucleases, endonucleases, polymerases and DNAzymes. Some exonucleases functioning on dsDNA could degrade one chain from the overhang ends to release another target chain into next cycle.418 Endonucleases, as a class of broadly used nucleases, had the functions of cleaving phosphodiester bond in DNA chains, where some DNA nicking endonucleases could specifically recognize and cut DNA sequences. In a branched DNA-based nucleic acid detection, DNA nicking endonucleases acted on specific cleavage sites, making DNA strands cleave into two pieces and release recycled target for amplification.419,420 This endonuclease-assisted target recycling strategy based on branched DNA could be developed to detect disease-related fusion gene or bacteria.370,421 Some restriction endonucleases worked on specific sequences, which limited the target detection of arbitrary sequences. To overcome the limitation, Wang et al. put forward a universal detection
AK |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. XXXX, XXX, XXX−XXX |

Chemical Reviews |
pubs.acs.org/CR |
Review |
Figure 40. Branched DNA-based materials for protein detection. (A) Illustration of DNA-based protein detection system formed by the combination of IgG antibodies (left), universal adapter (middle), and reporter molecules (right). (B) Multiple protein detection using universal adapter. Adapted with permission from ref 75. Copyright 2013, American Chemical Society.
strategy for di erent sequences of nucleic acid based on a restriction endonuclease.422 This strategy relied on a kind of Y- shaped structure composed of methylene blue (MB)-labeled capture probe with restriction site, assistant probe, and target DNA. With the digestion of restriction endonuclease, Y-shaped structure became unstable and dissociated and then released target DNA for circulation. Meanwhile, the MB-labeled partial sequences were away from gold electrode, resulting in obvious electrochemical signal changes. In addition to exonucleases and endonucleases, polymerases were also common tool enzymes for the manipulation of DNA sequences. Polymerases catalyzed the synthesis of DNA chains from four nucleotides using parental DNA as a template. The newly formed DNA chains occupied the binding sites of target DNA, thereby releasing target DNA. Xue et al. constructed an autonomously continues cycle process involving in target miRNA as catalysts and DNA polymerases as drivers (Figure 38C).369 Intracellular target miRNA promoted the strand displacement reactions of three hairpin DNA to achieve in situ imaging of miRNA. Compared with the target-induced linear nanostructures, the formed branched DNA exhibited a significant increasing in fluorescence intensity (Figure 38D). Liu et al. developed a one-pot cascade polymerization strategy for ultrasensitive detection of target DNA, in which target-induced Y-DNA probes, polymerasemediated target recycling and exonuclease-mediated cleavage signal amplification endowed it with highly sensitivity as low as 28.2 fM.382 Emerging as the catalytic agents, DNAzymes have received considerable attentions in molecular catalysis, computing circuits, biosensing and clinical diagnostics.423−427 Compared with traditional proteinases, DNAzymes had unique advantages including designability, easy operation, convenient condition, and low cost.428,429 DNAzyme contained a catalytic core flanked by separated recognition domains, which functioned to cleave specific substrates with the aid of cofactors.430,431 Zhang et al. employed a dual amplification strategy, including DNAzyme-based target recycling and signal transformation, in an ECL biosensor based on resonance energy transfer of molecule dye and quantum dots.365 Target miRNA as fuel triggered the assembly of hairpin chains to obtain Y-DNA containing Pb2+-binding cleavage sites, and then miRNA was released with the action of DNAzyme for the next cycle. At the same time, the released reporter DNA enabled the conformation conversion of DNA tweezer from “OFF” to “ON” state,
generating high e ciency of resonance energy transfer for the ultrasensitive detection of miRNA.
4.1.4. Multiplexed Diagnostics. By virtue of structural anisotropicity, branched DNA nanostructures possessed the capability of simultaneously detecting multiple molecules. When branched DNA was conjugated with expected fluorescence color codes, namely DNA nanobarcodes, they could be used to di erentiate miRNA expression level and cancer cells types according to in situ imaging of di erent cytosolic miRNA.432 Luo group created dendrimer-like DNA-based fluorescence- intensity-encoded DNA nanobarcodes, which were used for the multiplexed detection of pathogen DNA (Figure 39).73 Each DNA nanobarcode, conjugated to a molecular probe and two fluorescent dyes on the outermost layer, could be controlled in a specific color based on di erent red/green ratios of fluorescent dyes (Figure 39A). They immobilized the biotin-modified capture probes on the avidin-functionalized microbeads, for subsequent specific capture of target DNA and signal di erentiation display with di erent microbeads (Figure 39B). Using predictable fluorescence and specific hybridization of pathogen DNA and probes, these nanobarcodes were successfully utilized for rapid and high-sensitive detection of several pathogen DNA simultaneously via fluorescence microscopy, dot blotting, and flow cytometry (Figure 39C).
The conceptual design of DNA nanobarcodes provided the opportunities for the diagnosis of multiple protein. Tran et al. developed a versatile branched DNA-based protein detection strategy using a protein-DNA hybrid molecule called universal adapter.75 The universal adapter consisted of a universal protein binding moiety for integrating IgG antibodies and a DNA binding moiety for specifically combining with reporter molecules, generating a library of primary IgG antibodies (Figure 40A). This strategy successfully used a series of Y- shaped DNA nanobarcodes with di erent color ratio of red and green fluorophores to simultaneously detect three target proteins through dot blot technology (Figure 40B). In addition, quantum dots and horseradish peroxidase enzyme in replace of DNA nanobarcodes were also used as reporter molecules to detect proteins. The traditional protein diagnosis relied on the rational selection of available primary/secondary antibody pairs, which impeded the development of most antibody-based detection methods. This system not only circumvented the
AL |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. XXXX, XXX, XXX−XXX |
Chemical Reviews |
pubs.acs.org/CR |
Review |
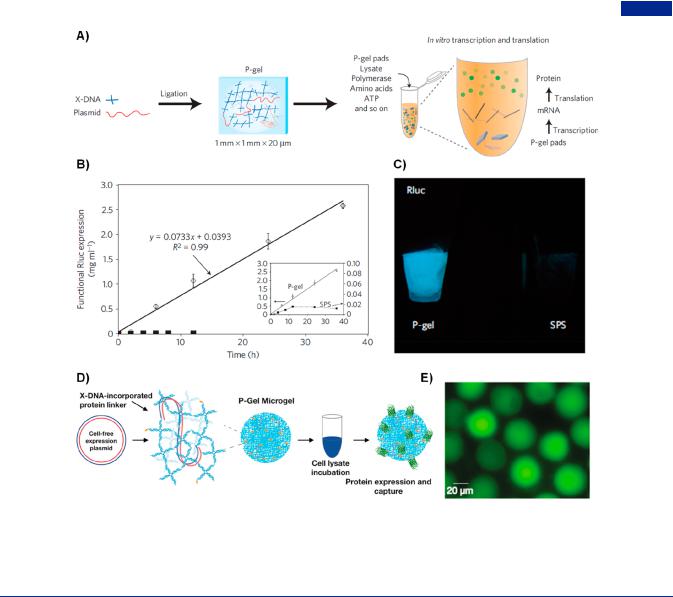
Figure 41. Cell-free protein producing gels (P-gel). (A) Formation of P-gel through enzymatic cross-linking and cell-free expression system. (B) Time courses of protein (Renilla luciferase, Rluc) expression. Open diamonds and filled squares represented P-gel and solution phase systems (SPS), respectively. (C) Bioluminescence images of Rluc protein in P-gel and SPS. Adapted with permission from ref 438. Copyright 2009, Springer Nature.
(D) Illustration of the formation of P-gel microgel constructed from chemically cross-linking of X-DNA network and gene. (E) Fluorescence display of GFP produced from cell-free protein synthesis system in P-gel microgel. Adapted with permission from ref 434. Copyright 2016, American Chemical Society.
use of secondary antibodies but also provided robust detection for multiplexed proteins.
To sum up, branched DNA have several advantages in diagnosis as follows: (i) sequence-specific molecular recognition enables branched DNA to bind to specific nucleic acid, protein or other molecules, making detection highly selective;402 (ii) the inherited hyperbranched structures can serve as signal amplifiers for ultrasensitive detection; (iii) the feature of topology allows branched DNA to combine di erent target molecules simultaneously, providing a possible for multiple detection; (iv) the flexible operation and multifunctionalities of branched DNA make it suitable for various types of signal outputs; (v) the operability of branched DNA can realize a recyclable detection platform;433 (vi) the size adjustability, good biocompatibility and biodegradability of branched DNA-based nanomaterials make the possibility of real-time intracellular monitoring.
4.2. Protein Engineering
As important biological macromolecules, proteins played essential roles in diverse cellular processes. In general, traditional cell-based protein production systems had the limitations of labor intensive, time-consuming, and inability to express various kinds of proteins due to cytotoxicity and inclusion bodies, severely limiting their applications in biomedicine. Instead, cell-
free protein production system could overcome these short-
comings and was an alternative advantageous protein expression platform.39,434−437
In 2009, Luo and co-workers invented a novel DNA hydrogel (P-gel as mentioned above) for the first time, which had the capability of producing functional proteins.438 By simply incubating with cell lysates, the resultant P-gel constructed by plasmid gene and X-DNA linkers expressed functional proteins through in vitro transcription and translation (Figure 41A). The P-gel system provided a relatively crowding environment for gene protection, a higher local concentration and gene proximity e ect, enabling an extremely high e ciency and yield of protein production (Rluc as the model protein) in comparison to solution-based cell-free systems (Figure 41B,C). Remarkably, 16 di erent membrane and toxic proteins were produced in the P- gel, which demonstrated the universal potentials in protein production technology.439 Inspired by the P-gel system, an RNA-producing nanohydrogel termed as I-gel was developed for RNA interference (RNAi) system.440 The nanohydrogel containing X-DNA and small interference RNA (siRNA) expression plasmid served as a self-circulating factory for siRNA production and a vehicle, exhibiting a higher transcription and interference e ciency through measuring the
AM |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. XXXX, XXX, XXX−XXX |

Chemical Reviews |
pubs.acs.org/CR |
Review |
Figure 42. Cell-free protein production system constructed from AuNP@gene circuit for studying the e ect gene circuit compartment on protein production. (A) Schematic of the construction of AuNP@gene circuit. Two mercapto-modified branched DNA with function-related genes coanchored on the surface of AuNPs for the construction of gene circuit compartment. (B) Schematic illustration of T7 RNAP transfer behaviors in gene circuit compartment. Schematic (below) showing the grafting e ciency and average grafting density of di erent AuNPs@gene circuits with the increasing of AuNPs. Adapted with permission from ref 442. Copyright 2019, American Chemical Society.
expression of target protein. The high performance of RNAi system was attributed to the improved stability of plasmid in I- gel, the higher local concentration of plasmid and continuous production of siRNA. Luo group further introduced linear
plasmids and X-DNA as cross-linkers into the DNA microgel sca old (Figure 41D).438 To e ectively display the protein
expression, a small part of X-DNA was functionalized by maleimido-C3-NTA used to chelate nickel ion and further capture GFP. The DNA microgels formed in microfluidic devices not only achieved gene isolation for a direct phenotype− genotype connection but also locally concentrated the gene for a higher protein expression productivity (Figure 41E). After the treatment of psoralen, the chemically cross-linked microgels were resistant to the denaturing conditions (such as high temperature) while maintaining the ability of producing proteins. Interestingly, the microgels had a capability for mimicking the inherited connection between genotype (gene enrichment) and phenotype (protein display) via fluorescenceactivated cell sorting (FACS), which provided a new research approach for protein engineering. More importantly, the protein-producing hydrogel behaved as cell subdivision, synthetic proto-nuclei, which was coated by lipid bilayers to reproduce natural nucleus system.441 The artificially engineering nucleus imitated the transcriptional system, allowing for a higher polymerase turnover rate and protein expression.
Very recently, Guo et al. delicately coanchored Y-shaped DNA with function-related gene expression cassettes on the surface of AuNPs to fabricate a gene circuit compartment and
deeply explore the compartment e ect on protein production (Figure 42).442 The gene circuit was composed of T7 RNA
polymerase and enhanced green fluorescent protein (eGFP) expression cassettes for gene regulation and report, respectively. The two gene expression cassettes were separately integrated into Y-shaped DNA nanostructure by advantage of psoralenmediated branched PCR technology (Figure 42A); at the same time, the other two branches of Y-shaped DNA were modified with mercapto groups to stably immobilize on the surface of AuNPs. The integrated nanointerface gene circuit created a compartment for regulatory gene and reporter gene, therefore e ectively enhancing the transfer e ciency of regulatory
proteins, cascade gene expression yield and initial rate, as well as biochemical reaction e ciency in cell-free systems. In addition, the gene expression yield, initial expression rate, and lag time of gene circuit were also a ected by the grafting e ciency and average grafting density of AuNPs@gene in varied gene circuit compartment systems. The phenomenon was attributed to the di erent T7 RNAP transfer behaviors in free gene circuit and gene circuit compartment (Figure 42B). The constructed nanointerface model interpreted the e ect of gene circuit in specialized subcellular compartments on biochemical reaction from the perspective of molecular mechanism.
4.3.Drug and Gene Delivery
4.3.1.Drug Delivery. Chemotherapeutic drugs usually bound to DNA in noncovalent ways of intercalation, groove binding, and phosphate electrostatic binding, where intercala-
tion and groove binding had certain selectivity to the conformation of chemotherapeutic drugs.443,444 For example, doxorubicin (Dox) with anthracycline structure, a most common chemotherapy drug for clinical treatment, was able
to preferentially intercalate into adjacent bases in GC-rich dsDNA.445 Because of the selective binding of drugs and DNA,
DNA-based materials were considered as excellent carriers for drug delivery.446−448 At the same time, the multibranched structure of branched DNA provided abundant intercalating sites. Luo group developed a kind of size-tunable and
monodisperse DNA nanospheres constructed from X-DNA for drug delivery.449 The X-DNA was self-assembled from four constituent strands with the modification of primary amine group, used to conjugate with a kind of photoreactive groups, poly(ethylene glycol) acrylate. Furthermore, the X-DNA monomer rapidly polymerized into DNA nanospheres upon UV irradiation due to photo-cross-linking groups. The DNA nanospheres had properties of concentration-dependent size adjustability and good monodispersity. DNA nanospheres e ectively delivered Dox into cells.
Systemic toxicity and low utilization e ciency of chemotherapeutic drugs remained a major concern. The emergence of
DNA aptamers provided a desired solution due to the high a nity targeting to corresponding ligands on tumor cells.445,450 Aptamer-based DNA materials possessed the advantages of
AN |
https://dx.doi.org/10.1021/acs.chemrev.0c00294 |
|
Chem. Rev. XXXX, XXX, XXX−XXX |
